Navigating the Depths of RNA Genomics Data
Bioinformatics
Looking for inspiration on your next custom RNA genomics project? Explore our interactive ENCODE RBP-eCLIP reports for your RBPs of interest; read our bioinformatics eBlogs to understand the latest RNA genomics tools; and learn about Eclipsebio’s Lunar™ Data Analysis Portal to help tackle your analysis
Let’s Learn
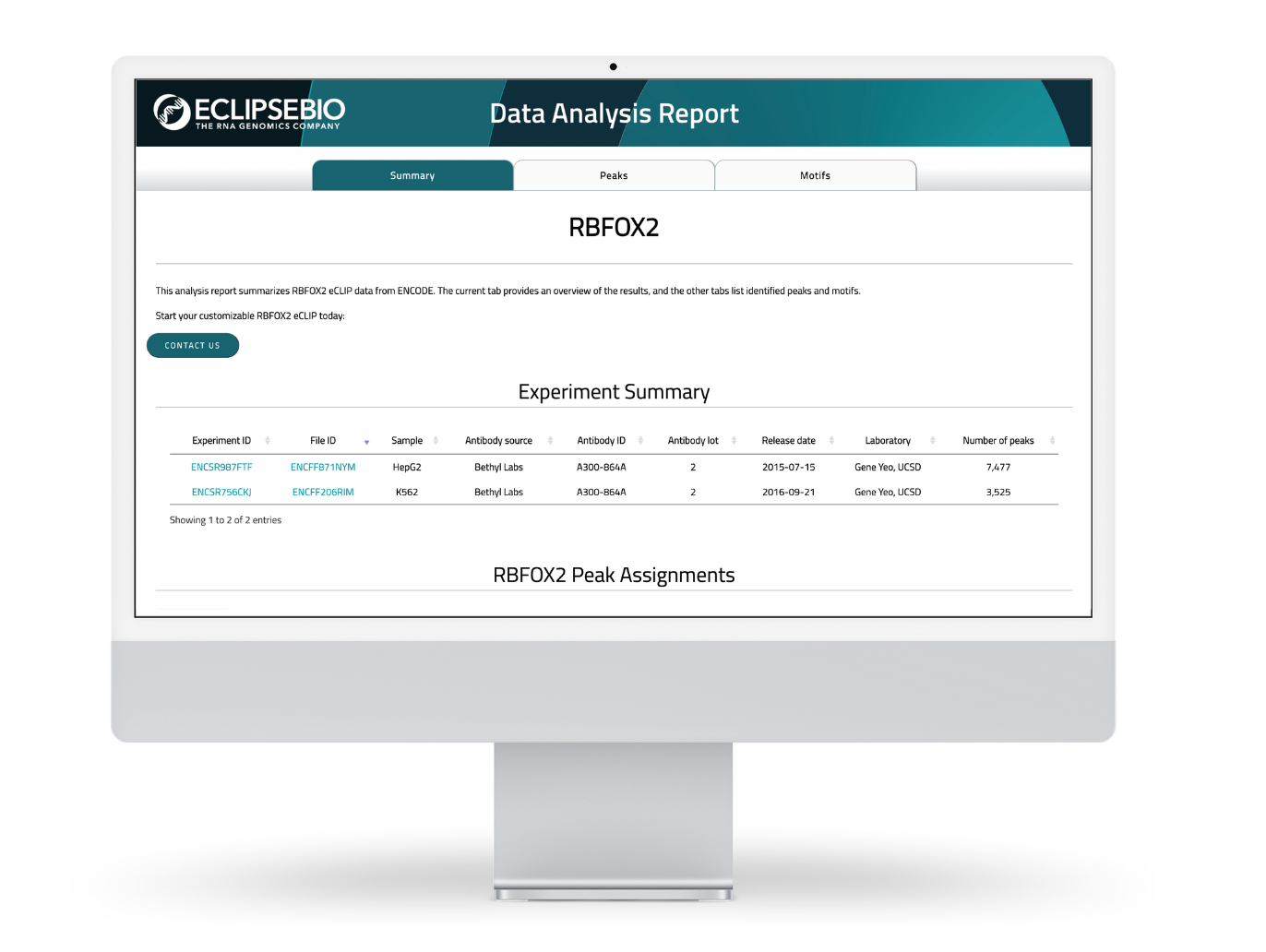
Download ENCODE datasets analyzed by Eclipsebio for your RBP of Interest
- Reports contain data for peaks and motifs
- Explore analyzed results from over 150 RBPs
- Discover motifs specific for your RBP of interest
- Compare your own results to ENCODE
Lunar Data Analysis Portal: Our Pipelines at your Fingertips
- All users can explore Eclipsebio demo RBP-eCLIP data deliverables in Lunar
- Directly download results or navigate through the data in Lunar’s integrated genome viewer
Read eBlogs by Expert Bioinformatics Scientists
- Interested in how we analyze our different technologies? Learn about new & internally developed bioinformatics tools for our latest RNA genomics technologies
- Gain insights into the different ways Eclipsebio examines data as well as general topics in the field of bioinformatics
eBlogs Written by RNA Experts
2024 | eBlogs
Discovering miRNA regulation with miR-eCLIP | eBlog
2024 | eBlogs
Successful mRNA therapeutic design: optimizing for stability and translation | eBlog
2024 | eBlogs
How to find stalled ribosomes | eBlog
2023 | eBlogs
2023: A Year-in-Review | eBlog
2023 | eBlogs
How to Use IGV: Part II | Bioinformatics eBlog
2023 | eBlogs
Thanksgiving 2023: Thankful for our customers and team | eBlog
2023 | eBlogs
Detection of Functional Small Open Reading Frames with Ribosome Profiling | eBlog
2023 | eBlogs
The Spooky Secrets of Identifying Protein:RNA Interactions | Halloween eBlog
2023 | eBlogs
How to Use IGV | Bioinformatics eBlog
2023 | eBlogs
RBP-eCLIP Motif Calling | Bioinformatics eBlog
2023 | eBlogs
RBP-eCLIP Peak Annotation | Bioinformatics eBlog
2023 | eBlogs
RBP-eCLIP Peak Calling | Bioinformatics eBlog
2023 | eBlogs
Stranded Libraries | Bioinformatics eBlog
2023 | eBlogs