During the process of translation, ribosomes move across mRNA sequences converting the information contained in codons into functional proteins. However, this process does not occur at a steady rate. Factors like the sequence of the mRNA and various cellular factors can cause ribosomes to pause, leading to stalls in translation at specific sites. These pauses can be critical for health or can be disruptive, leading to disease states or failed RNA therapeutics. In Fragile X Syndrome, the loss of an RNA-binding protein leads to decreased stalling contributing to the symptoms of the disease1. With mRNA vaccines, stalls due to RNA modifications can lead to ribosome frameshifting and mistranslation as seen with the SARS-Cov-2 vaccine2. Since the impact of stalls depends on the context of the RNA and cellular environment, it is critical to be able to locate and quantify ribosome pausing on a per-experiment basis.
Monitoring translation dynamics at the gene level can be accomplished by employing ribosome profiling technologies, such as Eclipsebio’s eRibo Pro service. Ribosome profiling, also called Ribo-Seq, allows researchers and therapeutic developers to obtain ribosome-protected footprints (RPFs), which are sequences of RNA that are undergoing active translation. By mapping where these footprints map to the transcriptome, the putative locations of ribosomes along a transcript can be identified. For instance, if the coverage near a start codon is significantly greater than the coverage along the length of the transcript, this can indicate that ribosomes are stalled or completely paused at or near the start codon. Identifying such events holds significant implications for biomedical research and the development of therapeutic mRNAs for vaccination and protein replacement. By pinpointing translational stalls or pause events, therapeutic and vaccine developers can enhance the translation efficiency of in vitro transcribed mRNAs leading to increased protein levels.
Several statistical techniques have been developed to distinguish putative stalls from normal rates of translation. These techniques range from traditional non-parametric tests to deep, recurrent neural networks. Regardless of the approach, putative pause events are characterized by the magnitude and position of their signals (e.g., the value of the statistic measuring the likelihood of a real pause event). At Eclipesebio, we have adopted a pause event detection analysis into our eRibo Pro service for ribosome profiling enabling robust identification of stalls.
In the first step of our method, a Kolmogorov-Smirnov (KS) test is used to determine if the mean coverage of RPFs along a transcript is significantly different between two groups of samples (e.g., treated versus untreated). The putative stall site is the codon position at which the mean coverage between conditions differs the most, indicating that one condition has accumulated RPF reads from paused ribosomes3. In a true pause event, most ribosomes will not progress past the stall site leading to a high number of reads prior to and a low number of reads after the identified site. To account for this, we only consider a given codon to be the site of ribosome stalling if the ratio of reads before to after the codon is significantly greater than one. Any identified stall site can then be visually examined in a genome browser to validate that a stall has occurred.
To assess our pause event detection approach, we performed an eRibo Pro experiment where we treated HepG2 cells with PF-846, a small molecule that induces translation inhibition at select genes including the cholesterol-related gene PCSK94. We performed a transcriptome-wide detection of pause events that were specific to PF-846 treated cells compared to untreated controls. Figure 1 illustrates the outcomes of this test. As anticipated, the transcripts exhibiting the most pronounced stalls were the same genes previously reported to be affected by PF-846, including PCSK9.
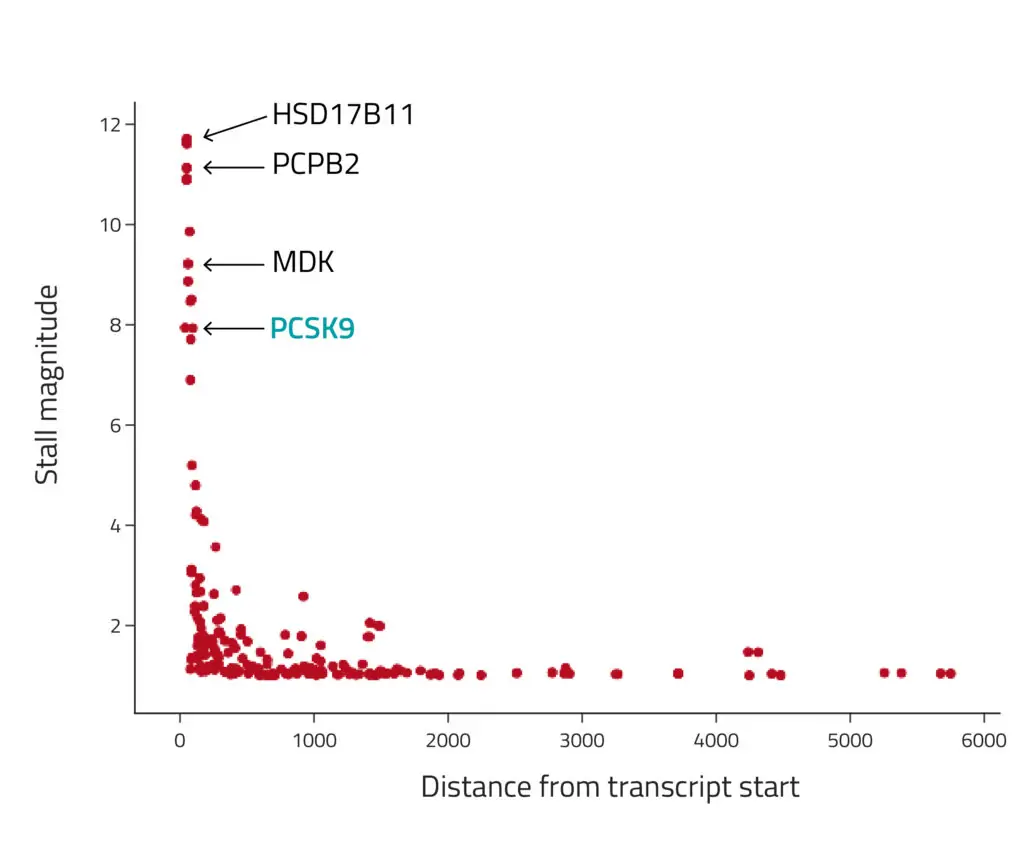
Figure 1: Detection of ribosome stalls after treatment with PF-846. The x-axis position denotes the stall positions, and the y-axis represents the fold change between coverage upstream and downstream of the stall position (i.e., the "stall magnitude"). The transcripts with the strongest stalls were the same genes previously reported to be affected by PF-846, including PCSK9.
On an individual gene level, we confirmed that treated samples exhibited a stall at the beginning of the transcript compared to the controls by visualizing reads along these transcripts in IGV, a tool for visualizing sequencing data. Notably, this effect is specific to the RPF portion of our ribosome profiling libraries. The RNA-Seq data, which reflects transcriptome abundance rather than ribosome association, is consistent in both conditions (Figure 2).
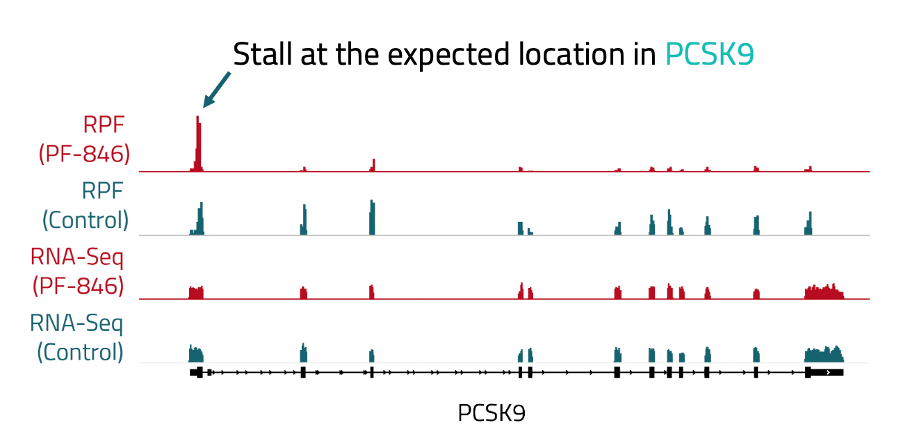
Figure 2: Bigwig tracks showing RPF and RNA-seq peaks along the PCSK9 gene. Tracks from libraries treated with PF-846 are highlighted in red. Tracks from control libraries are shown in blue. A bigger peak indicates more reads.
These results underscore the capability of ribosome profiling techniques to robustly identify where ribosomes stall. Eclipsebio's eRibo Pro service includes this analysis and all results are presented in interactive reports along with various other tests including codon usage, 5' loading ratio, and 3' read-through analyses. These analyses are instrumental to determine the impact of therapeutic or experimental conditions on translation and to assess the effective translation of in vitro transcribed mRNAs.
Contact Eclipsebio today to learn how eRibo Pro can help your research or therapeutic development program by identifying pauses in translation.
References
Latest eBlogs
Charting a New Era: How RNA Is Unlocking N‑of‑1 Cures
RNA is unlocking the ability to develop life‑saving therapies at unprecedented speed. The recent success with baby KJ demonstrates how RNA medicine can move a personalized treatment from diagnosis to clinic in less than a year.
The three pillars of AI in RNA biology: why data is the hardest to get right
Artificial intelligence (AI) is transforming how we approach RNA research and drug discovery. In this eBlog we review how data is one of the key pillars for the successful use of AI.
