As a young researcher, I was always told of the terrifying tales of CLIP-seq. It was described as a wicked protocol of radiolabeled probes and grueling long days, with no guarantee of producing a viable final library. Old lab lore told of times of failure regardless of how potent the antibody or expression level of the protein were. I would shiver in my lab coat thinking about consecutive failures and the Eppendorf tube graveyard that my bench was built on.

Thankfully, I never had to run the original CLIP-seq. With the growing importance of RNA binding proteins (RBPs) and their significant role in various diseases and pathways, it was clear that a better method of characterizing them must be found. Enhanced crosslinking and immunoprecipitation sequencing, or eCLIP-seq, was created to improve CLIP-seq methods and increase not only sensitivity but overall success of creating a final library. With a 1000-fold difference in library yields from CLIP-seq, eCLIP can robustly identify RBP binding sites transcriptome wide. Since its publication (Van Nostrand et al., 2016), eCLIP has been cited more than 700 times. Over 250 unique RBPs have been characterized by ENCODE. Currently, there are predicted to be over 2500 different RBPs, with the list of potential RBPs growing as prediction algorithms and AI become more powerful.
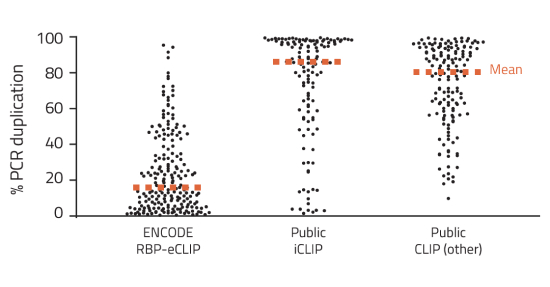
Despite eCLIP simplifying and improving CLIP-seq methods, eCLIP still sends shivers down researchers' spines. It is a 5–6-day protocol that is heavily antibody dependent and cell type or tissue type dependent. Optimization of antibodies can take weeks to months, and if there are no suitable antibodies, then epitope tags must be incorporated into the samples. Another monster under the bed is that tissues and cells can be difficult to collect. eCLIP requires ~20M crosslinked or 40 mg of cryoground tissues per sample but depending on the expression level of the protein in that sample type, sometimes double or triple that amount is needed for a usable library. These are all complications before the experiment begins, and without eCLIP expertise, many mistakes can happen during a 5-day protocol. It is for these reasons that the ghost of CLIP-seq still haunts eCLIP to this very day.
At Eclipsebio, we aren’t afraid of ghosts! Our RBP-eCLIP services can help you obtain meaningful data about your RBP of interest. RBP-eCLIP is a modified version of eCLIP that includes more quality checkpoints throughout the experiment. These QC points include RNA QC, which checks the quality and quantity of your RNA, and the biotin blot, which visualizes the overall RNA pull down from your target protein. This optimized method combined with the hands of our expert technicians and bioinformaticians can help you minimize the overall error and time taken to prepare and analyze your data. Included in these services are a step-by-step update of how your project is progressing, a full report of wet lab procedures, and an interactive HTML report that thoroughly outlines the results of your experiment.
Avoid the scares and tricks this spooky season and let Eclipsebio treat you with our RBP-eCLIP services!
Latest eBlogs
Charting a New Era: How RNA Is Unlocking N‑of‑1 Cures
RNA is unlocking the ability to develop life‑saving therapies at unprecedented speed. The recent success with baby KJ demonstrates how RNA medicine can move a personalized treatment from diagnosis to clinic in less than a year.
The three pillars of AI in RNA biology: why data is the hardest to get right
Artificial intelligence (AI) is transforming how we approach RNA research and drug discovery. In this eBlog we review how data is one of the key pillars for the successful use of AI.
