Technology Overview
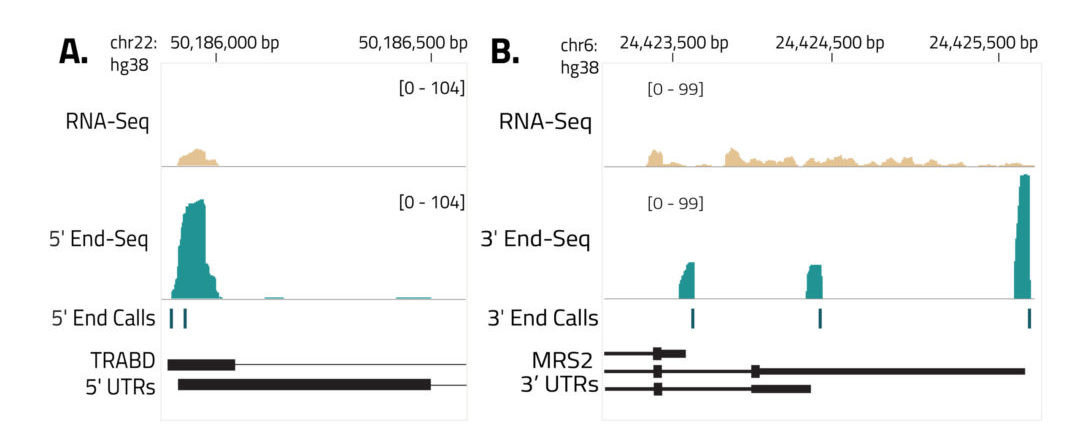
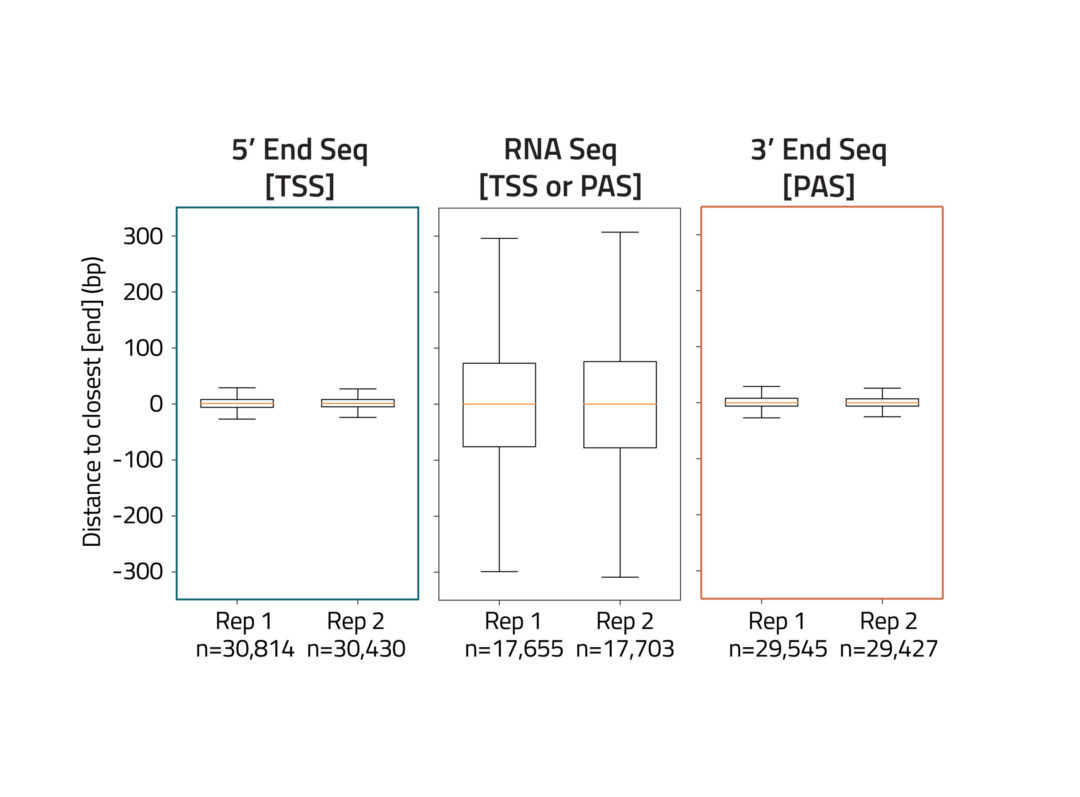
Precise mapping of transcription start sites (TSS) and polyadenylation sites (PAS) is critical to understand gene regulation and transcript diversity. Alternate TSS and PAS usage can either include or exclude key regulatory elements found in the untranslated regions (UTRs) at the ends of transcripts, ultimately having a profound impact on the molecular and cellular physiology of cells. Regulatory elements in 5′ and 3′ UTRs have been implicated in human disease and can function as therapeutic targets.
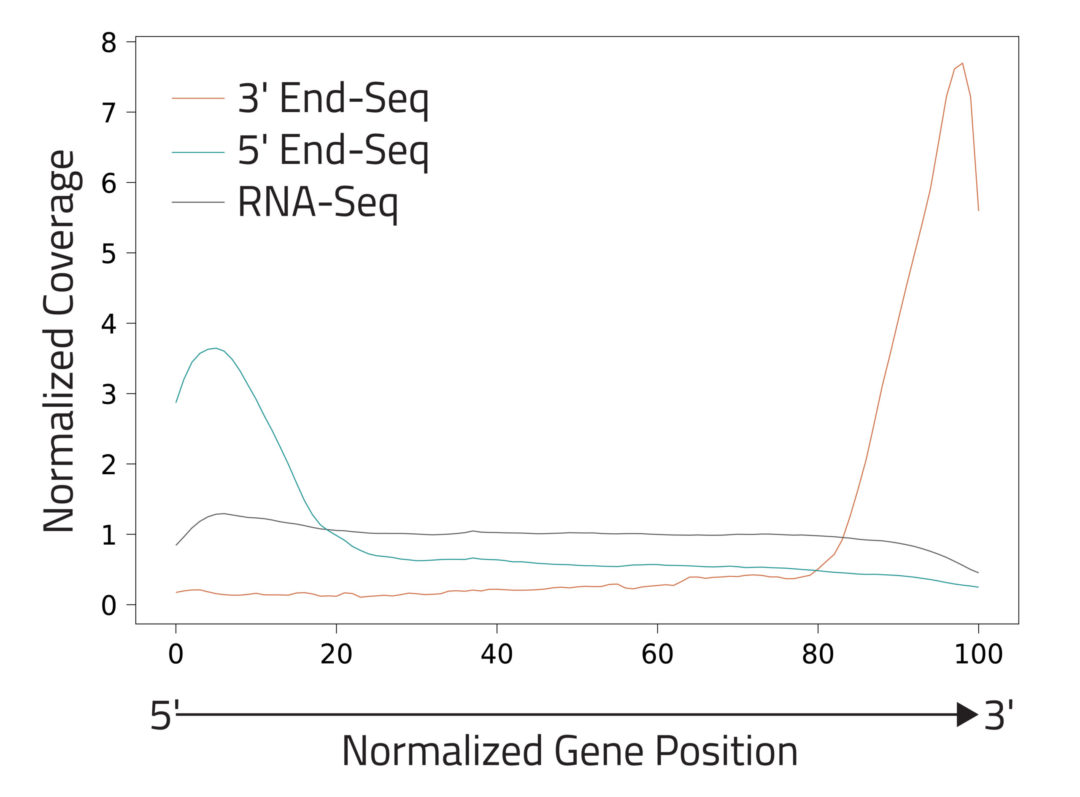
The End-Seq technology enables mapping of the 5′ ends and 3′ ends of transcripts by sequencing from the TSS and PAS, respectively, revealing the complete UTR landscape of expressed transcripts. 5′ and 3′ End-Seq enrich for transcript ends to detect known and novel TSS and PAS at single nucleotide resolution.