Overview
eSHAPE db-K562 is a deep sequencing database containing in vivo and in vitro SHAPE MaP data, as well as ∆SHAPE, the combination of in vivo and in vitro mutation rates, which can identify RNA positions that interact with proteins data from the K562 cell line.
The eSHAPE db-K562 is based on 6 technical replicates of polyA-selected RNA, sequenced at 125M reads each and is delivered as a data package for a single gene of interest or transcriptome-wide. Contact your Eclipsebio expert to see it check coverage of your gene of interest or to learn more.
BIOINFORMATICS
Our Comprehensive eSHAPE Per Gene Data Package
GENOME ALIGNMENTS | BAM
NAI treated and DMSO control reads that have been filtered of repetitive elements, aligned to the human reference genome (hg38), PCR deduplicated and are located within the start and end of the longest annotated transcript isoform for the gene of interest
eSHAPE REACTIVITY | BEDGRAPH
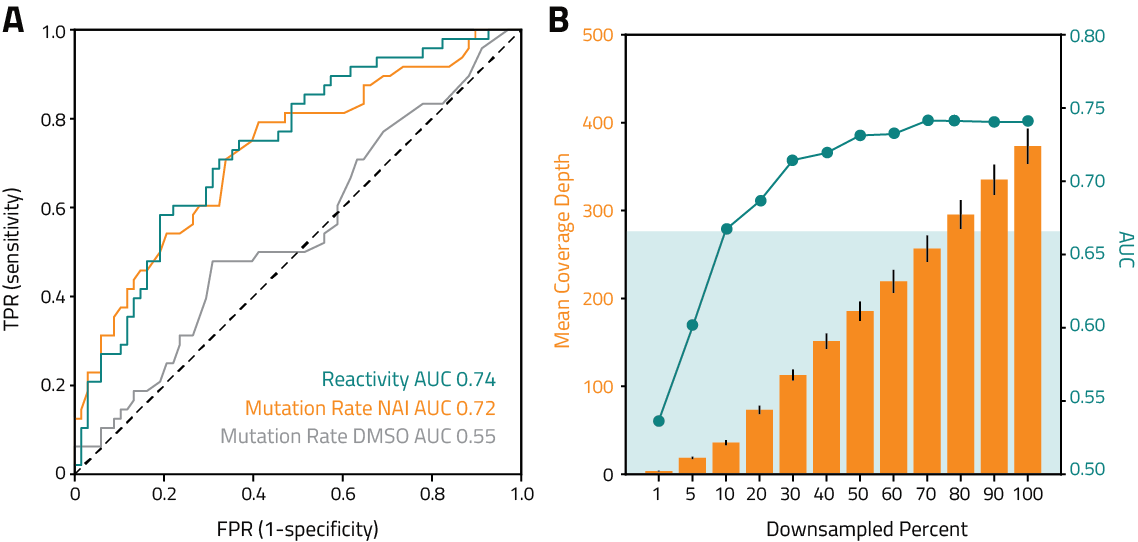
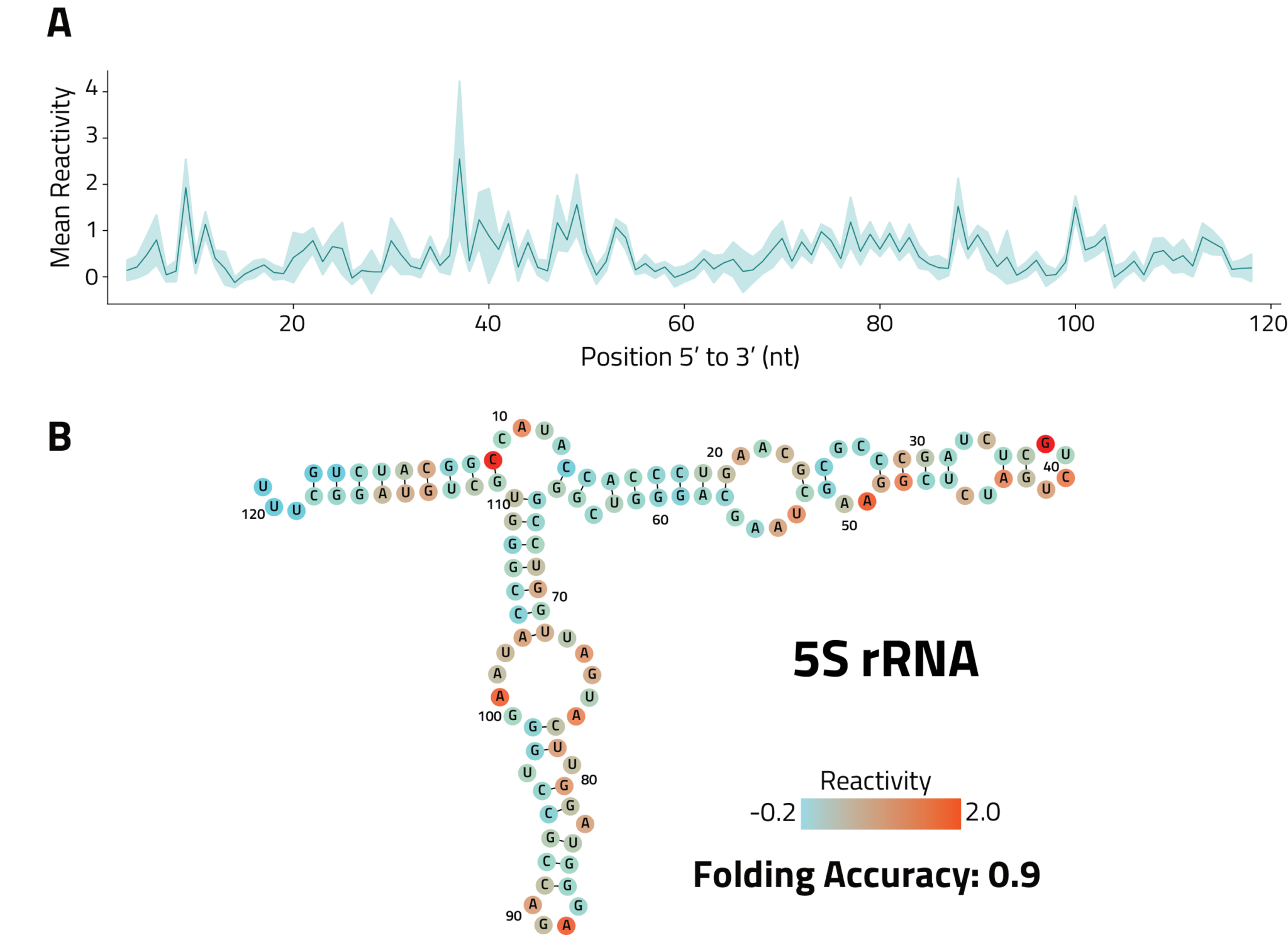
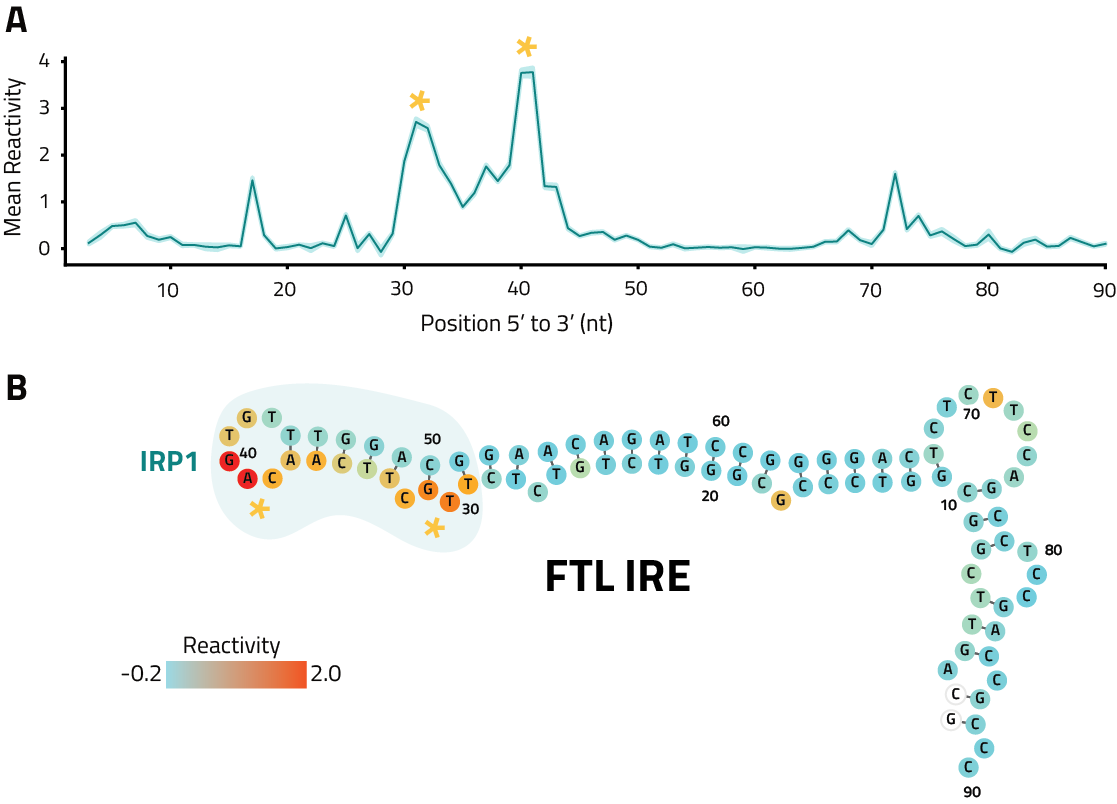
Raw reactivity scores at each position with >300x coverage within the start and end of the longest annotated transcript isoform for the gene of interest for visualization in a genome viewer such as IGV
PER TRANSCRIPT REACTIVITY | SHAPE
For each annotated transcript isoform of the gene of interest, IQR normalized reactivity scores at each position with >300x coverage with corresponding annotation sequence, for use as input into the RNAStructure algorithm to guide transcript fold predictions
GENE SUMMARY REPORT | HTML
Report of eSHAPE db-K562 results for the gene of interest with interactive tables and plots for publication ready graphics. Includes QC metrics, coverage, mutation rates and reactivities See Example Data Analysis Report