Choosing what model system to work with is one of the most important decisions a researcher interested in studying RNA biology will make. Across biological and pharmaceutical research, mammalian cell culture is one of the most commonly used model systems. In general, culturing mammalian cells is a scalable, flexible, and economical approach with important applications in basic research, drug screening, and the production of biologics. In this eBlog, we review cell culture and confluency and how they affect protein translation measurements.
Primary versus immortalized cell lines: choosing the right approach
Growing mammalian cells in culture might seem like a straightforward process, but there are many factors to assess to ensure reproducibility and achieve successful outcomes. Two particularly important considerations are the origin of cells and the capacity of those cells to divide. When considering what cells to culture, two major classifications exist: primary cells and immortalized cells. Primary cells are isolated directly from tissues or blood and tend to have no or limited proliferative ability after isolation. Consequently, they reach senescence relatively quickly. Primary cells are generally more difficult to culture but provide more direct insight into the physiology of source tissues, have relatively stable patterns of gene expression, and can contain heterogeneous mixtures of cell types. On the other hand, immortalized cells are highly proliferative and homogenous. The rest of this blog post will focus on these cell lines, which are also referred to as continuous. Unlike primary cells, immortalized cells maintain highly proliferative gene expression programs because they are typically derived from tumor sources and genetically modified or predisposed to divide indefinitely. Although we will be diving into the advantages of and best practices for culturing immortalized cell lines here, it is worth noting that there are several additional cell culture options that can provide unique benefits and biological insights including stem cells, organoid cultures, and hybridomas.
Measuring confluency is critical for effective cell culture
The health and proliferation of immortalized cells are highly sensitive to culture conditions. It is critical to maintain a consistent environment, including proper temperature, CO2 levels, and medium composition. Immortalized cells divide quickly, and therefore rapidly consume nutrients, produce waste, and occupy the physical space available in the dish or flask they are grown in. As a result, cells must be frequently diluted, typically in a process called passaging or splitting. A major advantage of immortalized cell lines is that they can be grown for many generations in the laboratory. However, cell lines cultured at high passages can show chromosomal duplications, rearrangements, or mutations, which can affect their morphology, proliferation, and general cell health. Therefore, documenting passages is an important practice and poor cell maintenance can negatively impact experimental outcomes. Furthermore, when not properly split, cell viability decreases.
To split cells correctly and consistently, researchers rely on a metric called confluency to dictate passaging schedule. Calculating the confluency of cells is the gold standard for monitoring their growth and knowing when they should be split. For adherent cell lines, confluency is defined as the percentage of the culture vessel surface occupied by cells. For instance, cells grown to 50% confluency will occupy 50% of the surface area of their dish or flask and are considered sub-confluent. Cells are considered fully confluent when 100% of the surface area is covered. In most circumstances, cells should be split before they become confluent. For suspension cells, which float freely in their growth medium, confluency measurements are conveyed by their density. To calculate confluency, one must take care to accurately count cells. Typically, cell counting requires techniques like trypan blue staining, loading of a hemocytometer, proper imaging with an appropriate microscope, or measurements using an automatic cell counter.
The confluency of cells is directly related to the growth phase they are in. When growing cells in culture, they typically progress through four phases: 1) lag, 2) log or exponential, 3) stationary, and 4) death (Figure 1).
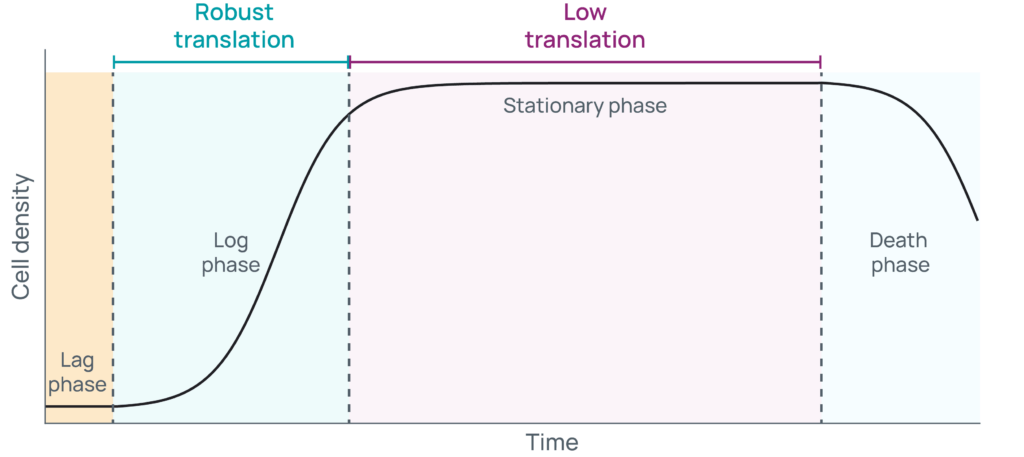
Typically, freshly thawed or split cells are plated at low sub-confluency and might undergo a brief period of latency as they acclimate to optimized, nutrient replete conditions. As cells transition from lag phase to log phase, they begin to express the RNAs and synthesize the proteins needed to rapidly divide. During this time, cells will grow exponentially and predictably, doubling in a specific unit of time. Cells cultured to a high-confluence state will next enter stationary phase, where death rates increase while growth rates decrease. For a time, those rates will be equal until waste accumulates and nutrients are depleted to the point where cell death dominates. Carefully monitoring the growth of cells is critical to know what phase they are in, which in turn provides several key advantages. To start, consistency in handling is a key component of reproducibility when generating biological replicates. Additionally, ensuring cells remain sub-confluent helps keep them growing, so they can continue to be passaged and used in multiple experiments in the future.
Confluency is a key determinant of gene expression and translation
The confluency of cells also has a direct impact on the RNAs they express. In turn, RNA is a critical player in many important biological processes. At Eclipsebio, we offer a variety of approaches to better characterize the functions of RNAs expressed in a researcher’s cell line of interest. To achieve success in the cell culture hood, one must understand the relationship that exists between confluency, growth state, and RNA content in their chosen cell line. One biological process where growth state and RNA abundance are particularly important is translation, a process where mRNAs are decoded by ribosomes which synthesize encoded proteins. Cells that are sub-confluent have the resources needed to rapidly produce mRNAs and translate them, and therefore tend to have high translation rates. Sequencing-based assays to study protein synthesis, such as our eRibo Pro ribosome profiling workflow, benefit from translationally active input material. Therefore, we recommend that cells are harvested for ribosome profiling at a sub-confluent state, at 70% confluency or below, to maximize the likelihood of capturing mRNA footprints from active ribosomes. In general, we encourage researchers to carefully count their immortalized cells, track confluency metrics, monitor passage number, and avoid stationary and death phases to achieve successful outcomes. Proper culturing techniques, when combined with the proper input of RNA required for an assay, are the best ways to ensure success when creating samples from immortalized cell lines.
Conclusion
If you would like to learn more about ribosome profiling, head over to our blog on identifying stalled ribosomes. Finally, reach out to our team if you’d like to learn more about how eRibo Pro or any of our other cell-compatible approaches work to reveal the RNA biology in happening in your favorite cell line.
References
Latest eBlogs
The power of pairing RNA design and analytics
Discover how closed-loop RNA development pairs design and sequencing to accelerate better therapeutics.
Achieving regulatory requirements by measuring IVT RNA integrity
As RNA therapeutics and regulatory requirements advance, validation methods for IVT RNA integrity are advancing as well.
