This blog was originally posted by Terrain Bio, now part of Eclipsebio.
What happens when your star performer mysteriously stops working?
This month, we witnessed an interesting phenomenon in our lab: our highest-performing mRNA construct—one that expressed 3x better than industry standards—was breaking in half. Not randomly and not from nucleases! It was breaking at a specific, predictable point. Read on to learn how we solved the mystery.
The unexpected breakdown: What we saw under the fragment analyzer
We recently tested several GFP mRNA sequences, including some that showed really great expression—up to 3x better than industry standards. The winner was crystal clear: the sequence with the highest expression, a clean fragment analysis trace, ready for action.
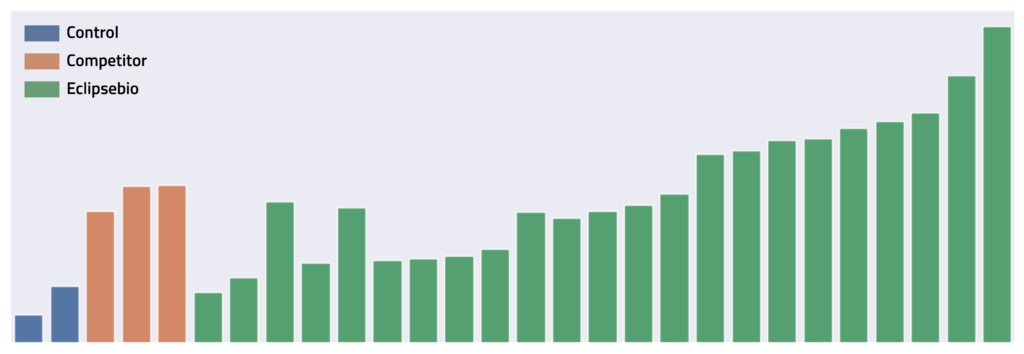
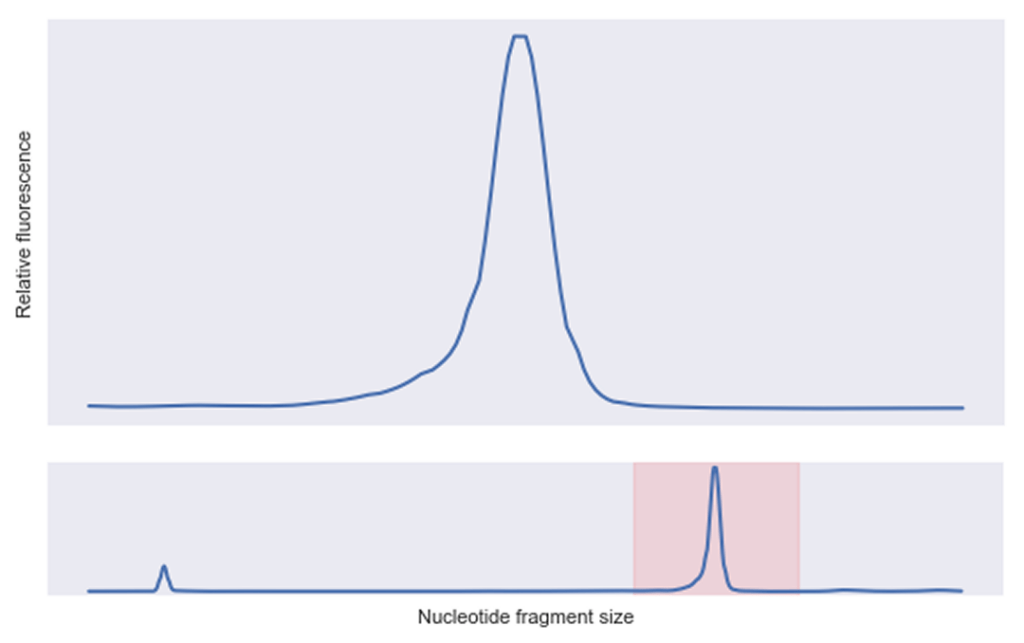
Fast forward three months: the same high-performing sequence was pulled from our -80°C freezer for follow-up studies. What we found surprised us: our sequence had degraded into a distinct fragment.
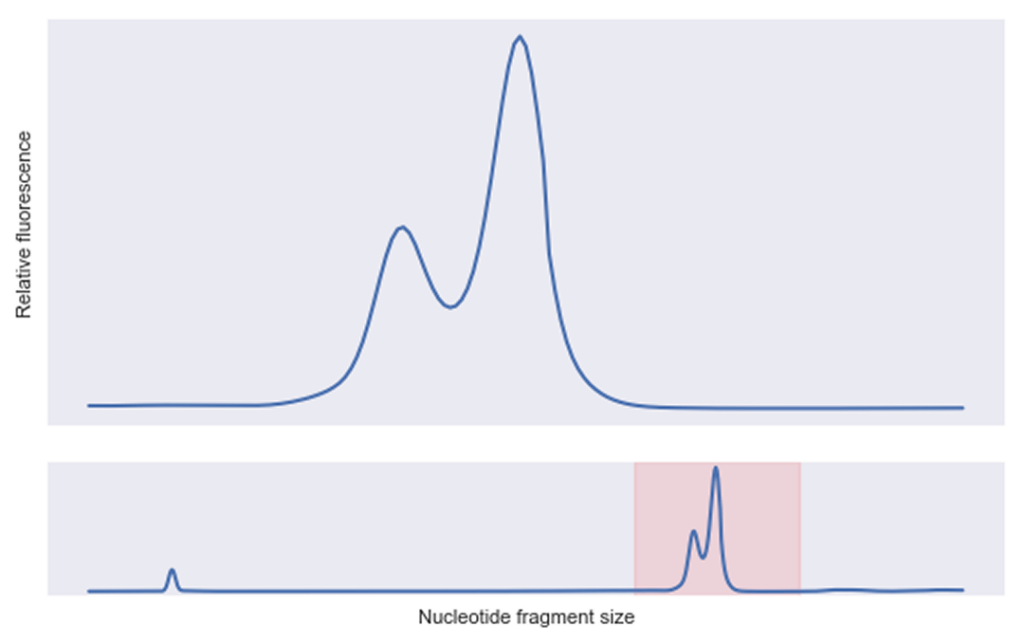
This pattern immediately caught our attention — it wasn't random degradation or RNase contamination. We were seeing a clean break at a specific site, resulting in the clear bimodal pattern in our electropherogram. Our team recognized this as a telltale sign of hydrolysis.
Beyond RNase: A fundamental chemical vulnerability
When mRNA mysteriously degrades, the default culprit is usually nucleases. So, we initially wondered if we had a nuclease contamination issue in our lab. But when we tested other constructs stored under identical conditions, most showed perfect integrity. This pointed us toward a sequence-specific vulnerability: hydrolysis.
Our team recognized this as a telltale sign of hydrolysis.
Hydrolysis can happen at specific hotspots determined by the sequence itself and its secondary structure. These vulnerable points can degrade over time even in -80°C storage. Through computational analysis, we've found that these hydrolysis hotspots aren't random: they’re largely predictable based on sequence and thus avoidable. We're now incorporating this insight into our partners’ projects.
The problem? Most sequence design tools optimize for a single metric: expression. They don't account for stability factors like hydrolysis susceptibility. What good is high expression if your construct falls apart before you can use it?
Multifactorial design: Eclipsebio’s specialty
At Eclipsebio, we've learned that successful mRNA sequences need optimization across multiple dimensions. Expression matters, but so does stability. A sequence that expresses brilliantly but degrades rapidly isn't viable for the lengthy timelines of therapeutic development (or in patients’ bodies).
By considering multiple factors simultaneously during design, we can identify sequences that perform well AND remain stable over time. This discovery reinforces Eclipsebio's multifactorial design+build approach.
We're continuing to investigate these hydrolysis patterns across diverse RNA sequences and building an ever-improving prediction model. It's just one example of how our team's scientific curiosity leads to better mRNA designs for our partners.
Are you seeing unexplained degradation patterns in your mRNA? Your sequence design might be hiding hydrolysis hotspots. We’d love to help you scrub those out and improve your mRNA. Chat with our team today.
Latest eBlogs
The power of pairing RNA design and analytics
Discover how closed-loop RNA development pairs design and sequencing to accelerate better therapeutics.
Achieving regulatory requirements by measuring IVT RNA integrity
As RNA therapeutics and regulatory requirements advance, validation methods for IVT RNA integrity are advancing as well.
