Key highlights
- Traditional RNA sequencing methods rely on short reads, which can obscure or lose important information.
- Direct RNA sequencing with Oxford Nanopore preserves full-length RNA molecules and their base modifications.
- This approach reveals key insights such as modification locations, isoform identification, and poly(A) tail length profiling.
- Direct RNA sequencing is also a powerful tool for RNA therapeutic quality control, enabling confirmation of sequence identity, detection of byproducts, and monitoring of modifications.
Introduction
Next-generation sequencing (NGS) technology has unlocked countless insights into the fundamentals of biology and made tangible improvements to human health. However, when it comes to understanding RNA, most NGS techniques rely on indirect sequencing of RNA molecules. Typically, RNA is fragmented, converted into DNA, amplified, and then sequenced as short reads. This approach has many advantages, including scalability and cost-effectiveness, but these steps can also introduce biases and obscure information contained in the original RNA molecules.
An introduction to direct RNA sequencing
Platforms such as Oxford Nanopore preserve RNA features that short-read sequencing loses by offering the ability to sequence RNA molecules directly. This technology works by threading full-length RNA through a nanopore. As nucleotides pass through the pore, changes in electrical current are recorded. These fluctuations are then decoded using basecalling algorithms to determine the sequence of the original RNA molecule as it passes through the pore.
Unlike short-read methods, this technique does not require steps such as fragmentation or reverse transcription. As a result, it maintains critical layers of information about the starting RNA molecules. Sequencing and basecalling on Oxford Nanopore’s platform enable single-nucleotide resolution of base modifications, poly(A) tail length determination, and isoform identification. Simply put, the power of this approach comes from the ability to take intact RNAs and profile them from their 5’ start to their 3’ end without first manipulating or altering them.
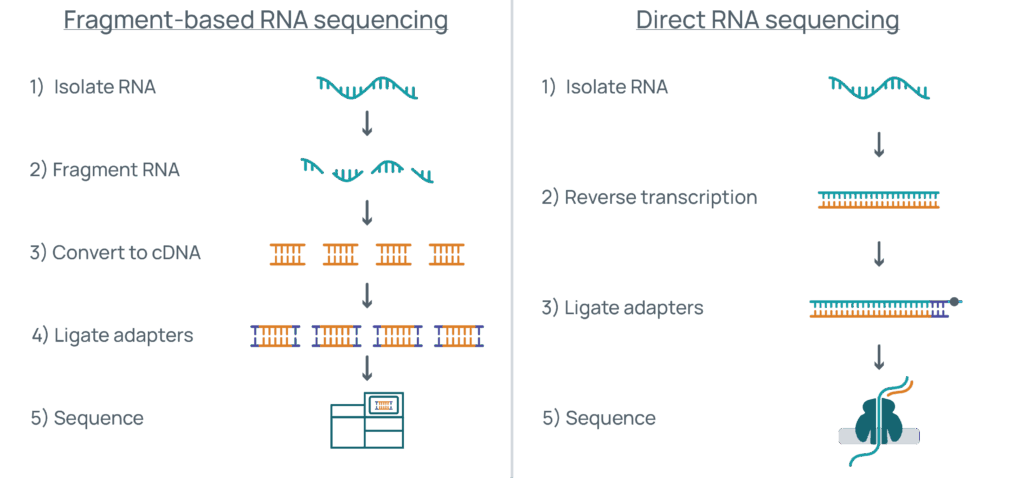
On the left, a traditional fragment-based workflow (e.g., Illumina) requires RNA fragmentation, conversion to cDNA, adapter ligation, and short-read sequencing. On the right, direct RNA sequencing preserves full-length RNAs. After isolation and reverse transcription to stabilize the RNA, a motor protein–linked adapter guides the molecule through a nanopore, where changes in electrical current reveal the RNA sequence.
Direct RNA sequencing in basic research
When profiling the transcriptome, these advantages allow researchers to explore aspects of RNA biology that are difficult to uncover with short-read sequencing. For example, a recent study used direct RNA sequencing to analyze how the transcriptome changed in response to knockdown of the m6A methyltransferase METTL3 in human cells1.
Researchers profiled and characterized features such as RNA abundance, poly(A) tail length, the impact of alternative splicing on isoform identity, and the presence of base modifications across transcripts. This approach revealed new insights into RNA biology, including how METTL3 depletion influences RNA regulation and the interplay between poly(A) tail length and RNA stability.
Direct RNA sequencing for therapeutic QC
The utility of direct RNA sequencing extends beyond transcriptome studies. It is also a powerful tool for quality control (QC) of in vitro–transcribed RNA molecules intended for therapeutic use. We have previously discussed the importance of therapeutic RNA characterization and the features critical for designing successful mRNA medicines. Proper characterization is a multi-step process, beginning with informed design, continuing through evaluation of synthesis, and extending to efficacy testing and off-target profiling.
Direct RNA sequencing is especially relevant for QC after the production phase of this process. Developers of RNA medicines must confirm they are producing high-quality RNA, and long-read sequencing enables multiple assessments in a single experiment:
- Sequence identity: Detecting mutations such as substitutions, insertions, deletions, or rearrangements.
- Poly(A) tails: Measuring length and distribution, which affect stability and translation efficiency.
- Molecule integrity: Determining whether RNA is full-length or truncated.
- Base modifications: Confirming incorporation of modified nucleotides such as pseudouridine, which influence stability and immune activation.
- Byproduct detection: Identifying unintended transcriptional byproducts.
Importantly, a single round of direct RNA sequencing via Oxford Nanopore can provide information on all of these features simultaneously. This reduces processing steps, improves QC efficiency, supports consistency, and enables reliable monitoring of batch-to-batch variability. For example, Gunter et al. recently demonstrated how nanopore long-read sequencing (VAX-seq) can be used to simultaneously assess sequence identity, integrity, length, purity, and modification status in mRNA vaccine constructs2. These considerations are critical for RNA medicines, where minimizing heterogeneity ensures reproducible dosing and patient safety.
Conclusion
Although direct RNA sequencing is powerful, it remains limited in certain aspects, such as throughput and accuracy, which make it a complement rather than a replacement for short-read technology. As the technology evolves, however, the insights it provides will only become more valuable.
Currently, RNA sequencing on the Oxford Nanopore platform offers an exciting way to examine RNA in its native state. By preserving full-length molecules, base modifications, and poly(A) tails, it provides insights into RNA biology that short-read methods alone cannot capture. For RNA therapeutics, from mRNA vaccines to emerging modalities, this approach adds a critical layer of quality control, ensuring that products are intact, properly transcribed, accurately modified, and consistent across production runs.
Such rigorous QC can bolster regulatory standards and help ensure that therapeutic RNAs consistently meet the highest quality benchmarks. As the field of RNA medicine continues to expand, integrating direct RNA sequencing alongside other advanced profiling methods will be key to accelerating discovery, improving design, and delivering safe, effective therapies to patients.
Whether you are advancing basic research or developing RNA therapeutics, our direct RNA sequencing services can help. Reach out to our team to learn more.
References
Latest eBlogs
The power of pairing RNA design and analytics
Discover how closed-loop RNA development pairs design and sequencing to accelerate better therapeutics.
Achieving regulatory requirements by measuring IVT RNA integrity
As RNA therapeutics and regulatory requirements advance, validation methods for IVT RNA integrity are advancing as well.
