Key Highlights
- RNA molecules fold into complex secondary and higher-order structures.
- RNA secondary structure plays critical roles in RNA biology including stability and the rate of translation.
- RNA-based vaccines and therapies can be designed to have stable structures, although this can reduce the rate of translation.
- The accessibility of binding sites affects the efficacy of small molecule, siRNA, and ASO therapies.
- Direct profiling of RNA structure in the cellular context is critical for the effective design of RNA-based and RNA-targeting medicines.
Introduction
RNA is a dynamic molecule that folds into complex secondary and tertiary structures, and these higher-order structures play critical roles in its stability and activity. As a result, this makes measuring RNA structure critical for the effective design of RNA-based medicines, such as vaccines, and RNA-targeting medicines, such as small molecule therapies. In this eBlog, we review key aspects of RNA secondary structure and discuss how the analysis of RNA secondary structure contributes to therapeutic development.
Understanding the Basics of RNA Structure
RNA can fold back on itself, allowing distant nucleotides to interact through intramolecular base pairing to form secondary structures such as loops and helices1,2. The base pairing that drives these secondary structures is typically dominated by Watson-Crick base pairing (adenine paired with uracil and guanine paired with cytosine), although wobble base pairing (such as guanine paired with uracil) can occur as well3. These secondary structures can interact with one another to form complex tertiary structures that create the three-dimensional conformation of the RNA.
There are several algorithms available to predict secondary structure in silico from the nucleotide sequence. Although the RNA sequence is a key determinant for structure, other factors play significant roles in the final conformation of the RNA including the rate of transcription, the rate of translation, and protein binding. The accuracy of structure prediction algorithms can be enhanced using experimental methods such as chemical probing with SHAPE-MaP and SHARC1,2.
Key Features of RNA Structure
Although RNA structure is driven by base pair interactions, a diverse range of structures can form other than the prototypical helix associated with nucleotide base pairing. Some of the common structures that can form are shown in Figure 1 and include1,4,5:
- Helices or stems: a stretch of paired bases.
- Hairpin loop: a loop of unpaired bases at the end of a helix.
- External loop: a stretch of unpaired nucleotides at the end of an RNA molecule.
- Internal loop: a loop of unpaired nucleotides flanked by a helix on either side.
- Bulge loop: a one-sided loop, in other words there is a bulge of unpaired bases out of a helix while the other side and flanking regions are paired.
- Multi-branch loop or multiloop: multiple unpaired strands connected by helices.
- Pseudoknot: loops interacting with one another.
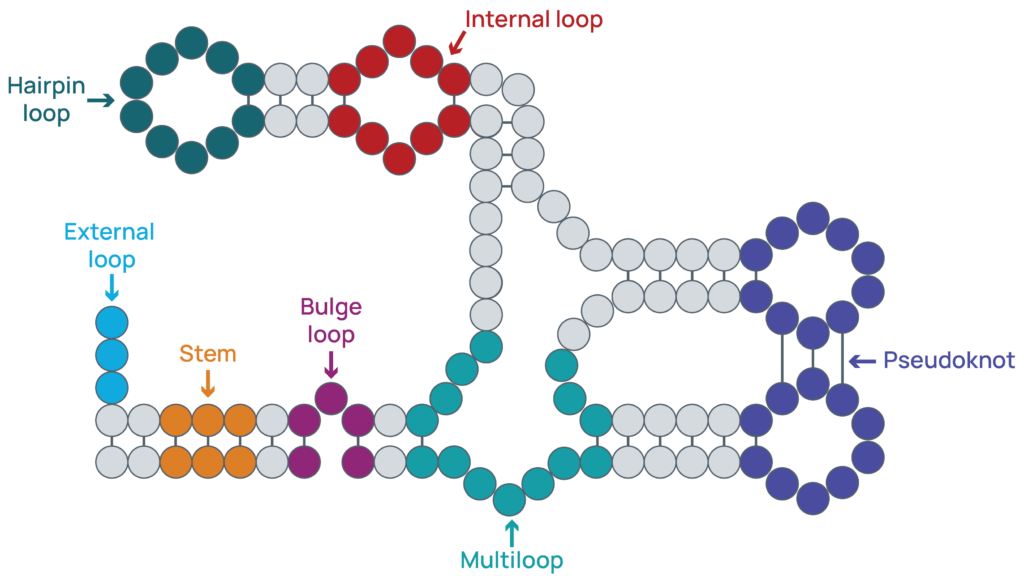
Figure 1: Common structural motifs. RNAs can fold into complex structures that regulate the stability and function of the RNA. Some of the common structures that can form are highlighted including stretches of paired bases (stems) and regions of unpaired bases (loops).
The Role of Structure in RNA Biology
RNA secondary structure is a critical determinant of multiple aspects of RNA function including:
- Stability: highly structured RNAs are less prone to degradation by ribonucleases or through hydrolysis6,7.
- Translation: the interplay between translation and RNA structure is an area of active research. It is typically reported that highly structured RNAs can reduce translation initiation and the overall rate of translation1,6,8. A decrease in structure and an increase in translation can lead to lower stability and RNA degradation, limiting the overall production of protein8,9.
- RNA-binding protein interactions: structure is a critical determinant of the ability of a protein to bind to and regulate RNA targets. Some RNA-binding proteins recognize specific secondary structures, such as stem-loop binding protein (SLBP, named for its preference for binding stem-loops in the 3’ UTR)1,10. For RNA-binding proteins that recognize specific sequences (motifs), these motifs need to be accessible for binding to occur11,12.
- miRNA binding: miRNAs bind to specific sequence motifs to guide RISC activity and the degradation of RNAs13. Like direct RNA-binding protein interactions, miRNA sites need to be accessible to enable binding and regulation by RISC14,15. For example, the binding of miR-221 and miR-222 to CDKN1B is enhanced when the RNA-binding protein PUM1 binds and induces a structural change in the 3’ UTR14.
- Splicing: splicing is a dynamic process that involves the recruitment of a large complex of RNA-binding proteins. RNA structure can affect splicing patterns by bringing splice sites into proximity, masking splice sites, or by masking repressor sites16,17.
RNA structure also plays significant roles in transcription18, ligand sensing with riboswitches19, and RNA localization20.
The Importance of RNA Structure in Therapeutics
With the importance of RNA structure across different RNA behaviors, it is not surprising that RNA structure plays a significant role in the development of RNA-based and RNA-targeting therapies. Below we review how RNA structure informs the design of several classes of RNA medicines.
Structural Stability and Translation of RNA Vaccines and Therapies
RNA-based vaccines and therapies need to be both translatable and stable. RNA secondary structure is a critical determinant in both aspects, however there is a balance between the two goals. Prior to entering a cell, highly structured RNAs are less prone to hydrolysis, making them more stable for long-term storage1,7. Within the cell, a highly structured RNA will maintain its stability, allowing for longer-lasting protein production6,9. The trade-off of this stability is that the rate of translation at any given moment will be lower than a less structured RNA due to decreases in translation initiation and elongation9,21. It remains unclear if it is more optimal to increase stability or translation, although the final decision will depend on the overall goals of the immunization or therapy.
Importantly, structure in specific regions can have different effects, requiring that drug developers examine a potential therapeutic holistically, with special focus on regions known to play significant roles, such as around the start codon and in the 5’ UTR6,9. RNA-binding proteins and other factors can alter RNA structure, so it is crucial that RNA structure is evaluated in a cellular context to obtain an accurate understanding of the drivers of therapeutic stability.
Small Molecule Drugs Targeting RNA Structures
Traditionally, small molecule drugs have been developed to target proteins. However, most proteins are considered undruggable, and many diseases have a non-protein component through the action of noncoding RNAs22,23. Due to these limitations, directly targeting RNAs with small molecules is a promising development for treating disease.
One approach for developing small molecules to regulate disease-related RNAs is to identify stable structures that form in vivo in disease states. Once these structures are identified, in silico studies can be performed to screen for potential binding and subsequent experimental validation24,25,26. As with RNA-based medicines, it is critical that structure is examined directly in the cell. Relying on in silico predictions of structure or in vitro measurements can often miss critical structural changes driven by cellular factors, which can affect therapeutic binding.
Small Oligonucleotide Drugs Targeting RNA Structures
As discussed above, the ability of miRNAs to bind their targets requires that the binding site be accessible for recognition by the RISC complex. The same accessibility requirements apply to small oligonucleotide drugs such as siRNAs and antisense oligonucleotides (ASOs), making it critical to examine RNA secondary structure during the development of an effective therapeutic.
Decades of research have found that structure is a key determinant of RNAi treatment effectiveness, though implementing structural information for siRNA design has been challenging27,28,29. The exact structural features associated with effective knockdown have varied across publications, but typically, structured regions such as helices are more preferred if part of the siRNA binding domain is accessible27,30. If the entire siRNA binding site is located within a helix or embedded in a complex structure that is difficult to unwind, therapeutic efficacy can decrease27,29. Surprisingly, an optimal binding site can include significant base pairing, which may reduce competition from RNA-binding proteins and miRNAs at the same site.
Like siRNAs, target site accessibility can improve the efficacy of ASOs. Reports of ideal target structures for ASOs vary, although unstructured regions and hairpins have typically been reported to be more effective31,32.
A major challenge for implementing RNA secondary structure into algorithms for small oligonucleotide design is that in silico predictions of structure are not always consistent with the actual conformation in the cell. The use of cellular-derived structures from experimentally-validated datasets can overcome the limitations of pure prediction approaches, improving the design of effective RNAi and ASO therapies29,33.
Conclusion
RNA secondary structure drives key aspects of RNA biology from transcription to protein generation and is a critical determinant of the effectiveness of RNA-based and RNA-targeting therapies. While, in silico predictions have helped in the design of different treatments, it is crucial that the conformation of RNAs in the cell is measured and used as a parameter for therapeutic design algorithms and AI models. Our eSHAPE assay and curated structure datasets can be used to train AI algorithms for therapeutic development or to validate the structure of a given RNA in different contexts, such as RNA vaccines in lipid nanoparticles (LNPs). To learn more about our support for designing and characterizing RNA medicines, contact us today.
References
- 1. Metkar et al.
- 2. Zhang et al.
- 3. Verani et al.
- 4. Wang et al.
- 5. Sato et al.
- 6. Leppek et al.
- 7. Wayment-Steele et al.
- 8. Mustoe et al.
- 9. Bicknell et al.
- 10. Corley et al.
- 11. Sanchez de Groot et al.
- 12. Connerty et al.
- 13. Iwakawa et al.
- 14. Kedde et al.
- 15. McGeary et al.
- 16. Shepard et al.
- 17. Saldi et al.
- 18. Georgakopoulous-Soares et al.
- 19. Garst et al.
- 20. Martin et al.
- 21. Neelagandan et al.
- 22. Neklesa et al.
- 23. Nemeth et al.
- 24. Child-Disney et al.
- 25. Tong et al.
- 26. Ribometrix
- 27. Schubert et al.
- 28. Piasecka et al.
- 29. Sagan et al.
- 30. Gredell et al.
- 31. Vickers et al.
- 32. Hörberg et al.
- 33. Hwang et al.
Latest eBlogs
The power of pairing RNA design and analytics
Discover how closed-loop RNA development pairs design and sequencing to accelerate better therapeutics.
Achieving regulatory requirements by measuring IVT RNA integrity
As RNA therapeutics and regulatory requirements advance, validation methods for IVT RNA integrity are advancing as well.
