- dsRNA is a rare in vitro transcription by-product that can affect RNA therapeutic translation and stability
- dsRNA can be produced by loopback events or from pieces of fragmented RNA
- Manufacturing optimization and/or purification can be used to reduce by-product levels
- Current detection methods are limited for quantification and optimization
Double-stranded RNA (dsRNA) consists of two RNA strands bound together. While RNAs are typically single-stranded, with the exception of secondary structures formed within the strand itself, dsRNA by-products can inadvertently form during the manufacturing of mRNA vaccines and therapies. dsRNA is typically associated with RNA viruses, the body often mistakes it for a viral infection, triggering an immune response. This response can lead to inflammation and other unwanted side effects as the immune system recognizes the paired bases through pattern recognition receptors, such as Toll-like receptors and MDA5. The presence of dsRNA is a major challenge for RNA-based therapeutics as it reduces protein production, decreases therapeutic stability, and can produce side effects.
Generation of dsRNA by-products
During an in vitro transcription (IVT) reaction, RNA polymerase generates a single-stranded RNA molecule called the run-off transcript. In relatively rare instances, dsRNA can be generated during this process. This can be through the RNA folding back on itself through complementary base pairing, called loopback, allowing the RNA polymerase to continue synthesizing past the poly(A) tail. Alternatively, pieces of fragmented RNA from abortive transcription or other copies of the RNA can bind to the run-off transcript, facilitating the re-engagement of the RNA polymerase and RNA synthesis (Figure 1).
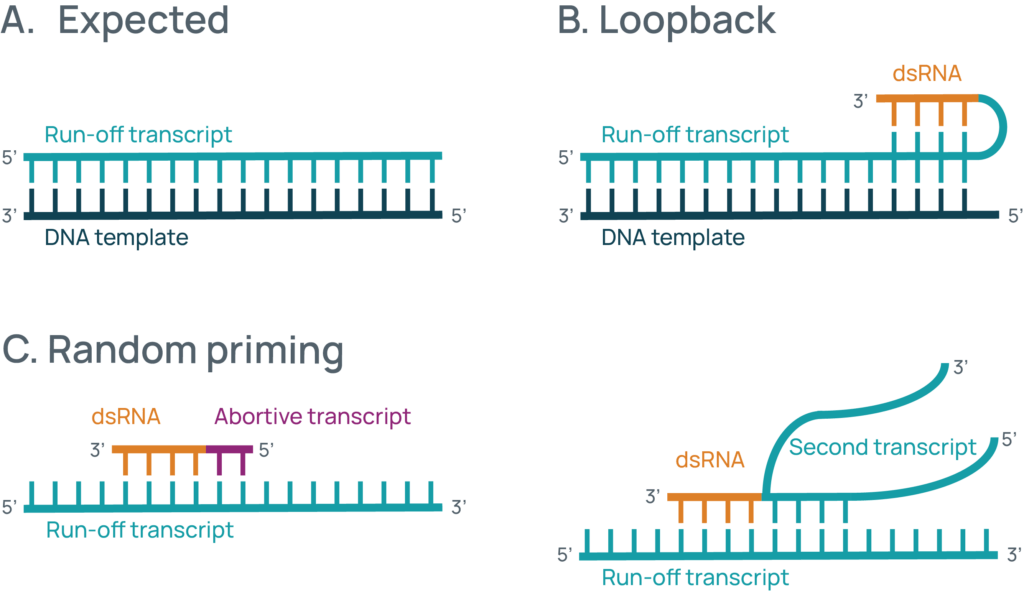
Figure 1: Typical sources of dsRNA. A) For most IVT reactions, a single run-off transcript is generated from the DNA template. B) In some cases, the RNA can loopback allowing dsRNA generation from the run-off transcript. C) dsRNA can also be generated from priming off other RNAs, either small transcripts generated from abortive transcription or from interactions between two run-off transcripts.
Limiting dsRNA in the final drug product
dsRNA by-products from IVT reactions are not a new phenomenon, and many strategies exist to reduce and remove their formation. Strategies for reducing its formation include conducting high temperature IVT reactions1, incorporating template encoded poly(A) tails1, maintaining low steady state levels of UTP2, using modified nucleotides such as pseudouridine3, and employing engineered RNA polymerases4. Methods for purifying IVT RNA include high ethanol cellulose chromatography5 and high-performance liquid chromatography (HPLC)6.
Quantifying dsRNA levels
Several methods for assessing dsRNA levels in RNA therapeutics already exist. These methods are typically based on standard biochemical methods such as polymerase chain reaction (PCR), northern blotting7, HPLC, reverse transcription quantitative PCR, fluorescence in-situ hybridization and mass spectrometry (MS)8. Although widely used, these methods are limited in their ability to detect dsRNA without pre-existing sequence information (PCR-based and hybridization methods), are typically low throughput, and require advanced infrastructure (HPLC and MS).
The most common methods, and the current USP recommendation for quantification in pre-clinical and clinical stage RNA medicines9, are antibody-based assays like dot blots or enzyme-linked immunosorbent assays (ELISA). Dot blot or ELISA assays are often used in combination with the J2-antibody10. The J2 antibody binds specifically to double-stranded regions, and does not bind to single-stranded RNA or DNA. However, the J2 antibody has several limitations such as sequence-dependency, cross-reactivity with RNA structures, minimum detection limits of approximately 30-40 nucleotide-long stretches of paired bases, and variability in antibody binding and signal detection, These limitations affect the reproducibility and reliability of quantification results, and prevent the identification of sequences and structures that generate dsRNA across the RNA therapeutic molecule.
Conclusion
Manufacturing by-products in RNA-based medicines can contribute to increased RNA degradation and limit RNA translation in the cell, negatively impacting the effectiveness of the therapeutic. The limited effectiveness of an RNA therapeutic may necessitate increased dosing, which can lead to increased side effects for the patient and cost for the manufacturer. Novel approaches for quantifying, characterizing, and optimizing dsRNA in RNA medicines are essential. An ideal approach should be non-sequence or size biased, capable of accurately quantifying levels regardless of nucleotide modification pattern, and be able to identify the source of dsRNA in the RNA to enable sequence-based optimization for reduced by-products.
Eclipsebio's eMERGE platform provides comprehensive approaches for quantifying and reducing manufacturing by-products. To learn more about our RNA therapeutic solutions, contact us today.
References
Latest eBlogs
Charting a New Era: How RNA Is Unlocking N‑of‑1 Cures
RNA is unlocking the ability to develop life‑saving therapies at unprecedented speed. The recent success with baby KJ demonstrates how RNA medicine can move a personalized treatment from diagnosis to clinic in less than a year.
The three pillars of AI in RNA biology: why data is the hardest to get right
Artificial intelligence (AI) is transforming how we approach RNA research and drug discovery. In this eBlog we review how data is one of the key pillars for the successful use of AI.
