miRNAs are small noncoding RNAs that regulate expression of their target transcripts. They are loaded into the RISC complex and target mRNAs based on perfect complementarity to their 5’ seed region (canonical binding) or through compensatory binding at the 3’ end (noncanonical binding)1. Their expression and activity is typically cell type or tissue dependent, limiting the ability of computational prediction methods to validate if a given miRNA is active in a given system. Understanding of binding and specific targets can help lead to advances in disease treatments, such as appropriate design of anti-miRs or avoiding competition with siRNAs.
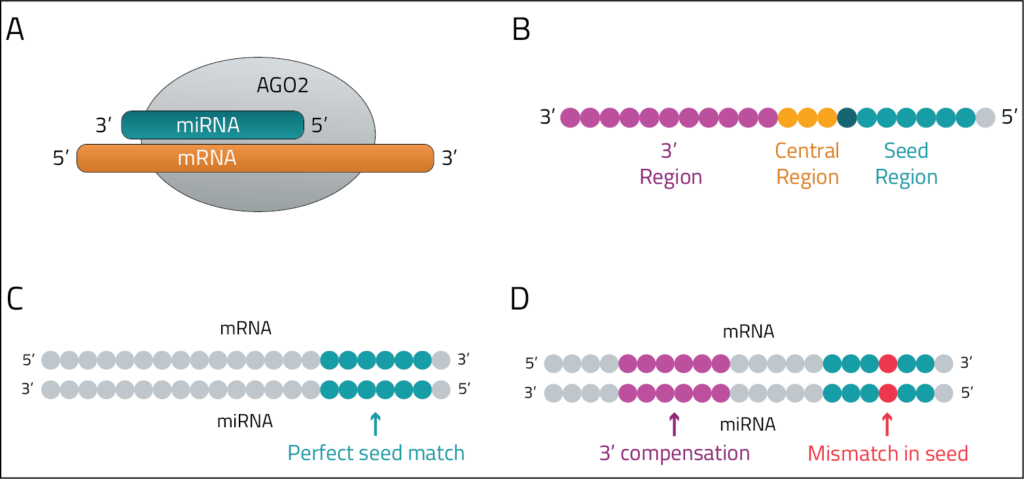
Figure 1: miRNAs bind to and regulate mRNAs. A. A miRNA binds in an antisense direction to its target mRNA within AGO2. B. miRNA regions including the seed region where canonical binding to the target mRNA occurs, the central region which can be involved in centered binding, and the 3' region involved in compensatory and supplementary binding. C. An example of canonical miRNA binding where a perfect match between the miRNA seed region and the mRNA leads to regulation. D. An example of noncanonical binding where a mismatch in the seed region is compensated for by binding in the 3' region.
miR-eCLIP reveals miRNA and siRNA binding
Our miR-eCLIP assay identifies direct miRNA-mRNA interactions transcriptome-wide, providing information on validated miRNA activity. The miR-eCLIP protocol begins with crosslinking of cells or cryoground tissue samples, followed by lysis and RNA fragmentation. RNA molecules bound to AGO2, a key member of RISC, are isolated using a AGO2-specific antibody, and an added ligation step forms chimeric molecules where each chimera contains both a miRNA and its mRNA target. We then perform library preparation, resulting in a next-generation sequencing library that contains validated miRNA-mRNA interaction data including chimeric reads. For our partners examining a particular miRNA or gene of interest, we can use probes to enrich for a specific sequence for increased coverage.
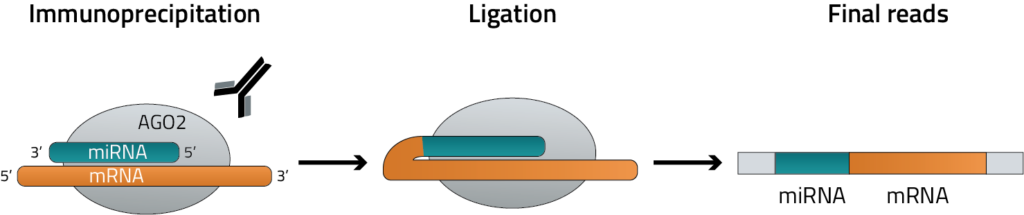
Figure 2: Overview of the miR-eCLIP assay. The RISC complex is crosslinked and immunoprecipitated with an antibody specific to AGO2. Reads are then chimerically ligated and next-generation sequencing libraries are generated where the final reads contain the miRNA sequence and the sequence of its mRNA target. This read structure then allows direct identification of each miRNA, its target, and where it is binding.
miR-eCLIP offers advantages over computational prediction methods such as TargetScan2. These methods typically focus on targets that contain a match to the seed region in the target’s 3’ UTR, where the seed region is typically defined as nucleotides 2-7 or 2-8 of the mature miRNA sequence. However, miRNAs can bind without full complementarity to the seed sequence or to other gene regions such as the coding exons. In addition, any output from one of these algorithms is a prediction and requires further experiments to validate that the binding occurs in a given cell type. With miR-eCLIP, we identify targets that bind due to complementarity to other portions of the miRNA as well as targets that do not contain any seed match. Due to the ligation between the miRNA and the mRNA, the assay provides direct information on miRNA activity, limiting the need for subsequent validation experiments.
miR-eCLIP can also be utilized to identify siRNA-mRNA interactions. miR-eCLIP +siRNA can identify siRNA off-targets and reveal differential targeting between multiple siRNA candidates. Analysis of siRNA chimeric reads can reveal alternative processing of siRNAs as well as indicate where the siRNA binds to its off-targets, regardless of whether the binding occurs in the canonical seed region. These features make miR-eCLIP +siRNA an invaluable tool for siRNA design and screening.
Transcriptome-wide profiling of miRNAs in the K562 cell line
To help support the RNA community, we offer free access to a miR-eCLIP dataset we generated from K562s, a widely-used lymphoblast cell line. The dataset is information-rich, containing over 7 million total chimeric reads and over 16,000 reproducible binding peaks.
To help with data exploration, we provide a summary HTML report that highlights abundant miRNAs in the K562 cell line as well as seed match presence in chimeric peaks. The report also includes visualization of enriched motifs within peaks and the seed matches associated with those motifs, as well as enriched gene ontology (GO) terms within the set of genes with bound miRNAs.
If you contact Eclipsebio, you can also download additional data files free of charge. The additional data package includes chimeric read BAM files, bigWig tracks, and a BED file of reproducible target peaks. These files can be visualized in a genome browser or analyzed further using bioinformatics tools. With this dataset, researchers and drug developers can focus on a particular gene of interest and see what miRNAs are targeting it or explore targets of specific miRNAs of interest.
Eclipsebio supports your research and drug development goals
At Eclipsebio, we are excited to help further your scientific discoveries and support siRNA and anti-miR drug development. Contact us today for more information about the miR-eCLIP K562 dataset or running a miR-eCLIP experiment in your sample type of choice.
References
Latest eBlogs
Reducing off-target effects in siRNA: expert tips for better efficacy
In this eBlog we discuss the mechanisms of siRNA off-target interactions and outlines strategies to minimize them.
RNA structure analysis for improved drug development
RNA secondary structure is a key determinant of RNA biology and therapeutic efficacy. In this eBlog, we review how RNA structure affects the development of RNA vaccines, siRNAs, and ASOs.
