RNA therapeutics such as RNA vaccines, antisense oligonucleotides (ASOs), siRNAs, and aptamers have already changed the face of medicine and their impact is only expected to increase in the years to come. Although RNA therapeutics are valued at billions of dollars1, there remain significant R&D costs and high risks of failure for any developed therapeutic2,3. Here we review why RNA therapeutic development can fail and ways to limit the risk.
#1) Off-target effects
Even the best designed therapeutic is expected to have some degree of off-target effects. These off-target effects can result from several causes such as off-target binding as seen with ASOs and siRNAs4,5 or through the activation of innate immunity as seen with Sirna-027 which was discontinued after the siRNA was discovered to activate TLR36. In the case of a siRNA therapeutic, off-target screening can be performed indirectly by computationally looking for matches between the siRNA sequence and genes but with the limitation that any identified off-target is a prediction and not validated. RNA-Seq can also be performed to look for decreases in gene expression, although it can be difficult to separate direct siRNA binding from indirect changes. With Eclipsebio's miR-eCLIP+ assay, the location of siRNA binding across the transcriptome can be directly measured reducing development risks and timelines.
Off-target binding of an siRNA was evaluated with miR-eCLIP+. We were able to find 90 validated off-target binding events on protein coding genes after siRNA treatment with no binding events detected in the absence of the siRNA.
# 2) Delivery challenges can limit efficacy
Another major challenge in applying an RNA therapeutic is delivering it to the target while maintaining efficacy. RNAs are prone to degradation, so different packaging systems have been developed such as lipid nanoparticles (LNPs) although these can act as double-edged swords by preventing degradation at the expense of higher clearance by the liver and kidneys7. These and other issues have often meant that therapeutics need to be delivered directly to the target tissue, as seen with Spinraza, an FDA-approved ASO drug that is directly injected into the fluid surrounding the spinal cord.
# 3) Protein binding affects therapeutic success
For many RNA therapeutics, RNA binding proteins are critical partners required for their activity. siRNAs require the action of the Argonaute family of RNA binding proteins to cleave the mRNA target8. Spinraza, an ASO drug for spinal muscular atrophy acts by regulating splicing, which is a complex process that involves more than 40 proteins9. Eteplirsen is an ASO that induces exon splicing, requiring the involvement of multiple RNA binding proteins10. Any RNA introduced into a cellular environment will have the potential to be regulated by one of the more than 2000 RNA binding proteins in the human genome11, which makes understanding their activity critical for therapeutic success. Eclipsebio’s RBP-eCLIP technology can be used to profile the binding of different RNA binding proteins, allowing drug developers to evaluate if a given protein is a critical effector of therapeutic efficacy or to identify proteins that could inhibit activity.
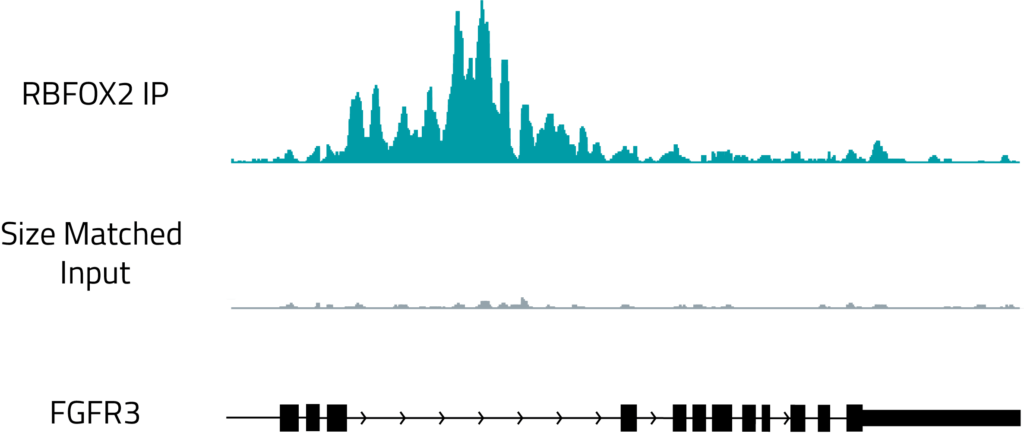
Genome browser track showing the binding of RBFOX2, a known regulator of splicing, to the intron of FGFR3. The top, teal track shows RBFOX2 binding and the gray track shows a size matched RNA control.
# 4) Low translation efficiency limits protein production
Many RNA therapeutics act by creating proteins (the spike protein of SARS-CoV-2), regulating what form of a protein is expressed (Spinraza's regulation of SMN1 splicing), or inhibiting translation (Fomivirsen12 and Tofersen’s regulation of SOD113). In cases where a protein product is being generated it is critical that translation occurs efficiently to produce therapeutic protein levels7. To determine if an RNA therapeutic has affected protein expression, different techniques can be used including mass spectrometry or Eclipsebio’s eRibo Pro assay which provides a snapshot view of both gene expression and protein translation in a single assay.
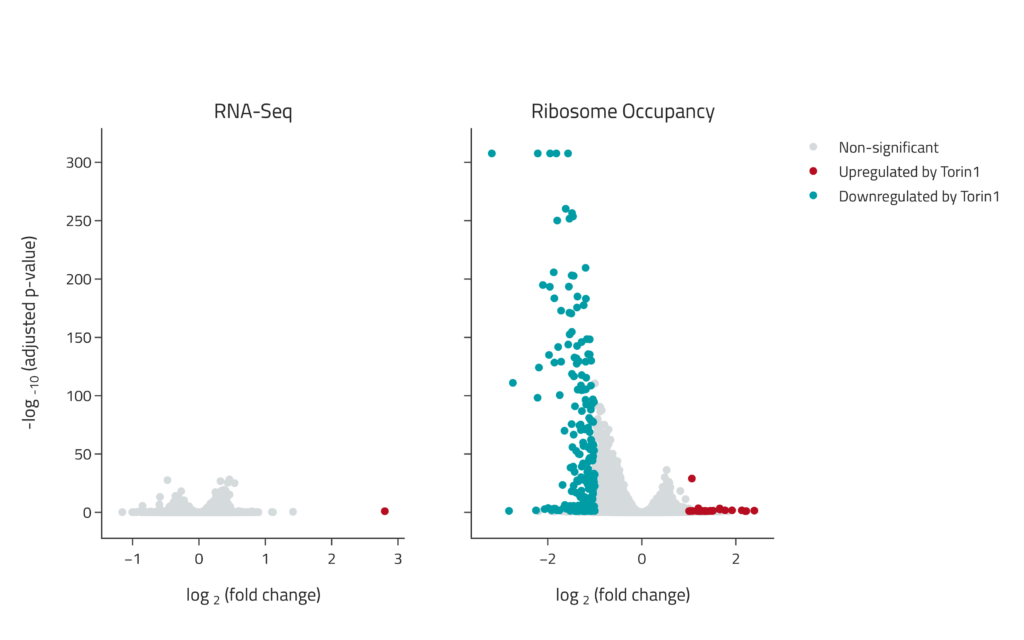
Samples were treated with the small molecule therapeutic Torin1 and changes in gene expression and translation were determined using eRibo Pro. The volcano plot on the left shows changes in gene expression (RNA-Seq) and the volcano plot on the right shows changes in translation (Ribosome Occupancy). Torin1 induced a specific suppression of translation with limited effects on expression.
# 5) RNA structure affects accessibility
RNAs are structured molecules with complex secondary and tertiary structure including stems, hairpins, bulges, and pseudoknots. This structure plays foundational roles in the level of protein translation14, therapeutic stability15, and the actions of ASOs16. Due to the important nature of RNA structure in therapeutic efficacy it is critical to have a robust and reproducible method for identifying RNA structure. RNA structure can be determined using NMR, X-ray crystallography or with Eclipsebio’s eSHAPE assay which provides a comprehensive SHAPE-based determination of RNA structure.
eSHAPE was performed on in vitro transcribed and in cellulo transcribed RNAs of the gene FTL. To determine where cellular interacting partners, such as proteins, are binding ∆SHAPE was calculated, which is the mutation rate in the in vitro eSHAPE experiment subtracted by the mutation rate in the in cellulo eSHAPE experiment. An increase in ∆SHAPE indicates that the base is more likely to be paired in the cellular environment, likely due to a protein binding or another cellular factor. The regions with higher ∆SHAPE on the shown RNA structure (asterisks) are known binding sites for the protein IRP1.
Conclusion
Developing an effective RNA therapeutic has a number of challenges, and at Eclipsebio we have developed a portfolio of solutions to ensure therapeutic success. Interested in learning how Eclipsebio can help you achieve your drug development goals? Contact us today.
References
Latest eBlogs
The power of pairing RNA design and analytics
Discover how closed-loop RNA development pairs design and sequencing to accelerate better therapeutics.
Achieving regulatory requirements by measuring IVT RNA integrity
As RNA therapeutics and regulatory requirements advance, validation methods for IVT RNA integrity are advancing as well.
