Key Highlights
- siRNAs are promising medicines for decreasing the expression of disease-causing genes.
- siRNAs can have off-target effects through miRNA-like binding.
- Asymmetric design, chemical modifications, pooling, and sequence design can reduce off-target effects.
- Validation of off-target binding is critical for RNAi success.
Introduction
Small interfering RNAs (siRNAs) hold immense promise as therapeutic agents, offering the potential to silence disease-causing genes with high specificity. Despite their proven success in the clinic, a significant concern surrounding RNAi medicines is the potential for off-target effects. siRNA therapies can unintentionally silence genes other than their intended target, leading to unforeseen consequences1,2. This eBlog will explore the issue of off-target effects and discuss design strategies to make them safer for your drug development programs.
Partial binding drives off-target activity
siRNAs are short, double-stranded RNA molecules that work by guiding the RNA-induced silencing complex (RISC) to cleave target mRNA molecules, effectively silencing the corresponding gene3,4. While the guide strand is designed to be complementary to a target mRNA, it can also potentially bind to and silence other mRNAs with partial sequence homology2.
This partial binding makes the siRNA behave similarly to microRNAs (miRNAs). miRNAs generally bind to the 3' untranslated regions (UTRs) of mRNAs with approximately a 6-7 base pair match in the seed region (Figure 1), leading to translational repression or degradation. siRNAs can similarly bind to off-target mRNAs through the seed region (bases 2-7/8 of the antisense strand). If there are mismatches in the seed region, the 3' region can compensate as well. This can lead to the downregulation of unintended genes, causing unpredictable and potentially harmful side effects2.
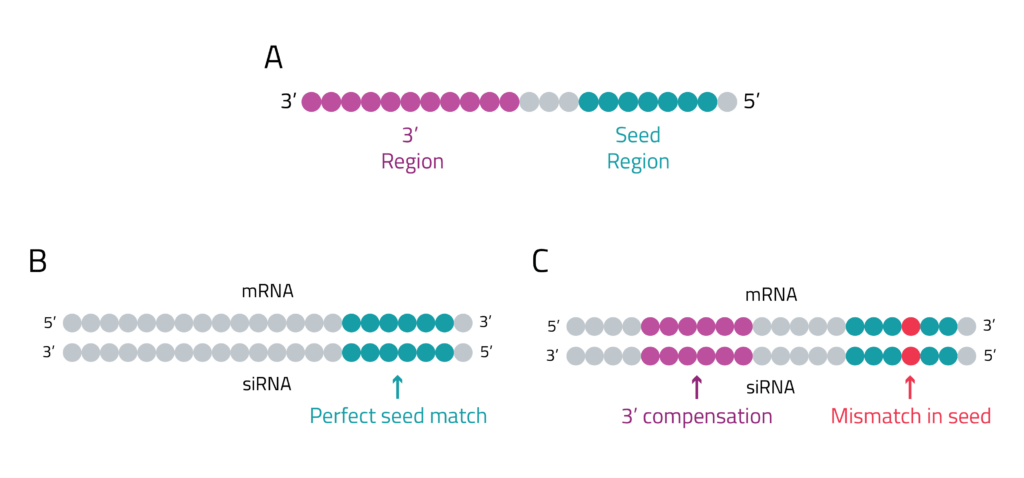
Figure 1: Off-target binding patterns. A. miRNAs have defined regions that can interact with targets including the seed (bases 2-7/8) and the 3' end, the same regions from siRNAs can also have distinct binding activities. B. siRNAs can bind like a traditional miRNA where the bases 2-7/8 act as a seed to drive binding. C. siRNAs can have mismatches in the first 8 bases but still have off-target binding due to compensation by binding at the 3' end.
To avoid off-targets, some combination of the following approaches is typically used:
- Asymmetric design
- Chemical modifications
- Pooling
- Optimized sequence design algorithms
Asymmetric sequence design can optimize RISC loading
The strand of the siRNA duplex that is less thermodynamically stable at its 5' end is preferentially loaded into the RISC complex6. This inherent bias can be exploited to promote the loading of the guide strand and minimize off-target effects caused by passenger strand loading. Strategies to promote guide strand loading include using chemical modifications to destabilize the 5' end of the passenger strand or designing therapies with a shorter passenger strand6,7. An alternative to traditional double-stranded siRNAs is the use of a single-stranded RNA (ss-siRNAs). These molecules can be designed to directly mimic the guide strand, bypassing the need for preferential strand loading8.
Chemical modifications can reduce off-target binding
Modifying siRNAs chemically can improve their stability, specificity, safety, and bioavailability. For example, 2'-O-methylation of the guide strand can decrease miRNA-like off-target effects and reduce immunogenicity without compromising the intended gene silencing effect9.
Common modifications include4:
- Phosphorothioate (PS) linkage: Replacing one of the non-bridging oxygen atoms in the phosphodiester bond with a sulfur atom is a common modification. PS modifications increase resistance to degradation by nucleases; however, excessive PS content can reduce the binding affinity between the siRNA and its target sequence and increase toxicity.
- 5′-(E)-vinyl phosphonate (5′-(E)-VP): This modification replaces the oxygen and carbon at the 5′-end of oligonucleotide with E-vinyl phosphonate moieties. This substitution increases stability, enhances binding to Argonaute-2 (a key component of RISC), and improves potency in vivo by increasing its accumulation and residence time in tissues. 5′-(E)-VP modifications have shown potential in single-stranded siRNA (ss-siRNA) applications, achieving favorable pharmacokinetic and pharmacodynamic profiles.
- Ribose modifications: Introducing modifications at the 2′ position of the ribose sugar (such as 2′-O-methyl, 2′-O-methoxyethyl, or 2′-fluoro) increases stability by protecting siRNAs from nuclease degradation, increases binding affinity to target mRNA by promoting a C3′-endo conformation in the ribose sugar, and reduces immunogenicity. However, it has been shown that excessive 2′- modification can significantly reduce RNAi activity.
Pooling limits off-target activity
Utilizing a pool of siRNAs that target various regions of the same mRNA can effectively reduce off-target effects while ensuring strong on-target silencing. By designing pools with distinct seed sequences for each siRNA, the effective concentration of any individual seed is reduced, thereby minimizing the risk of off-target silencing associated with seed sequence similarity2,10.
Algorithms and machine learning to reduce off-target binding
Computational tools such as BLAST can analyze siRNA sequences for homology with other genes, helping to identify and eliminate those at high risk for off-target effects during the design phase. Furthermore, advanced algorithms and machine learning models have been developed to predict efficacy and off-target potential by assessing sequence features, thermodynamic properties, and other relevant factors5,11,12.
Validation is required to ensure therapeutic success
Thorough experimental validation is crucial to identify and minimize off-target effects. Researchers often use multiple siRNAs targeting the same gene to confirm that observed phenotypes are due to on-target silencing and not off-target effects. Expression profiling (microarray analysis and RNA sequencing) is traditionally used as an indirect measure to assess global gene expression changes and identify potential off-target genes13,14.
Conclusion
The potential for off-target effects is a valid concern in the development of siRNA therapeutics. By understanding the mechanisms underlying these effects and employing the design strategies outlined above, drug developers can significantly minimize their occurrence and maximize their chances for successful drug development.
Continued advancements in sequence design, coupled with rigorous experimental validation, will pave the way for safer and more effective RNAi therapies for a wide range of diseases.
To learn more about how Eclipsebio can support the design and validation of siRNAs, visit our solutions for small oligonucleotides. Our assay provides a comprehensive and global view of siRNA and miRNA binding and can be utilized to enhance therapeutic design. We also offer datasets of structural accessibility and isoform detection across a variety of different cell lines and tissues, which can be used to train AI models for therapeutic design.
References
Latest eBlogs
The power of pairing RNA design and analytics
Discover how closed-loop RNA development pairs design and sequencing to accelerate better therapeutics.
Achieving regulatory requirements by measuring IVT RNA integrity
As RNA therapeutics and regulatory requirements advance, validation methods for IVT RNA integrity are advancing as well.
